Skip Navigation
Search
Helmut H. Strey, Ph.D.
Associate Professor
Research Focus

My lab "Micro- and Nanotechnologies for Quantitative Biology" is currently focused on three main areas of research:
- Cell-Cell variability: Through the development of a wide range of single-cell characterization techniques
it has become clear that mammalian cells of the same phenotype that are grown under
the same conditions, often exhibit significant cell- to-cell variability. Cellular
variability is often connected to understanding cancer recurrence and stem cell differentiation.
We are currently working on the development of a novel micropatterning technologies
that will allow us to observe clonal colonies that derive from a single cell while
allowing long-term repetitive observation of each individual cell. If successful,
our technologies enable measurements that will give us a unique insight into the cells
circuit architecture. From our experiments we anticipate discovering how tightly cellular
rate constants are regulated and to determine the relaxation times that describe how
fast fluctuations around the homeostatic value decay back to equilibrium.
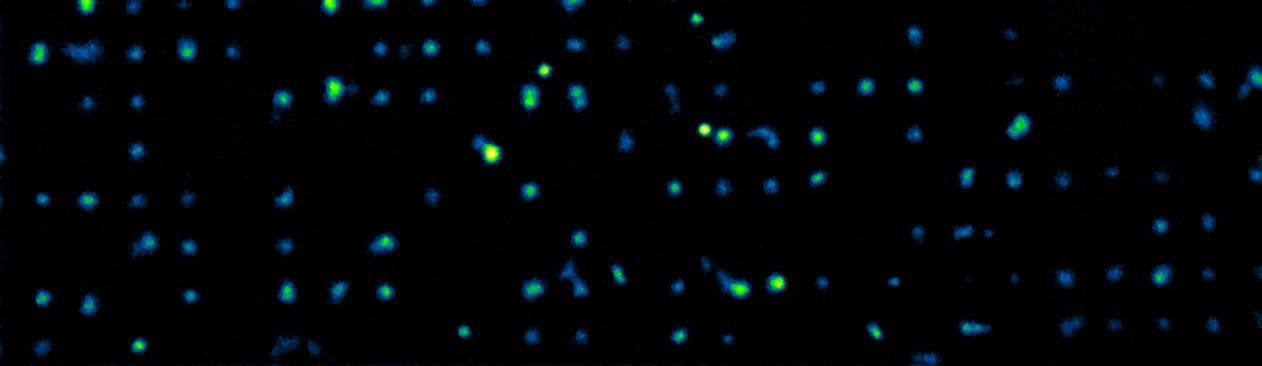
Single Cells on a Micropattern
- Cancer-on-a-chip: In recent years, the role of metabolism in cancer has become an area of intense interest.
In the 1920s, Otto Warburg suggested that cancer is a metabolic disease caused by
dysfunctional mitochondria after he found that tumors consume much more glucose than
other tissues and doing so by using the less efficient metabolic pathway of glycolysis
to produce ATP. Even though his original interpretation has been disproved, the Warburg
effect is still actively and intensely studied because it is linked to the emergence
of invasive and metastatic cancer. Most, but not all, invasive cancers exhibit a phenotype
that metabolizes glucose at a higher rate than non-invasive cells. At the same time
most invasive cell lines exhibit an increased utilization of the glycolysis even if
exposed to oxygen (aerobic glycolysis or Warburg effect). This is in contrast to “normal”
cells that only utilize glycolysis when exposed to hypoxia (oxygen deficiency), which
is also called the Pasteur effect. The Warburg effect has led to several hypotheses
to explain how aerobic glycolysis is advantageous to the fitness and survival of cancer
cells. We are currently working to create tumor-on-a-chip technology to grow well-defined
3D tumor microenvironments that can be exposed to defined nutrient stresses such as
differing oxygen and nutrient gradients. In addition, we will be able to determine
the metabolic phenotype of cancer cells with differing oncogenic changes and degrees
of aggressiveness. Our technology should be able to distinguish between competing
and synergistic hypotheses to address the question of the role of the TME on metabolic
reprogramming and in particular glycolysis.
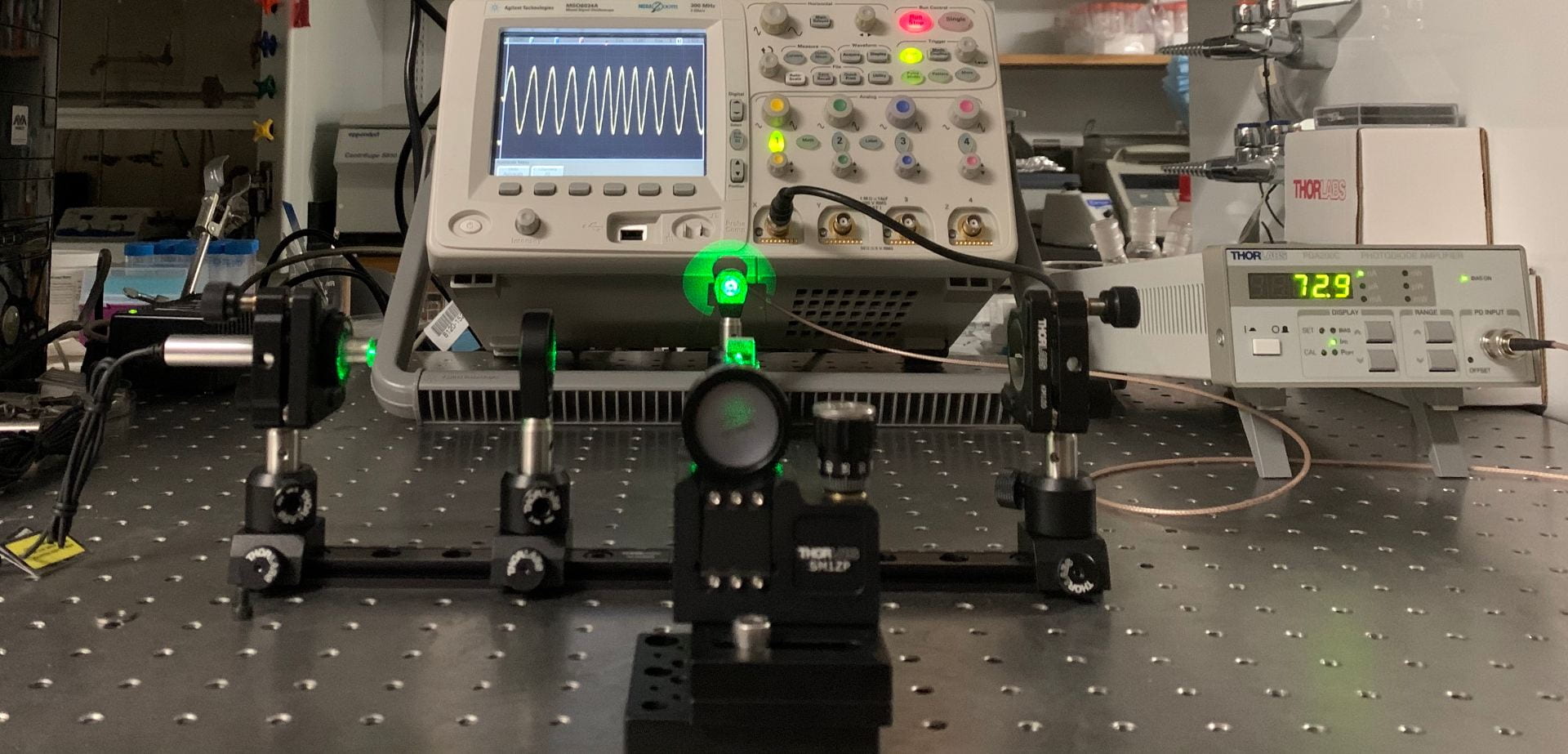
Development of an Optical Decoder for a Translation Stage
- Bayesian Analysis of time-series and Network discovery: Over the last few years, my group has developed several data analysis algorithms for time-series data. In particular, we have developed a comprehensive theory for the Ornstein-Uhlenbeck process, which describes a stochastic process that has a stationary solution (mean) and fluctuates around this mean through a Wiener process (random walk). Our maximum-likelihood solution results in the probability distributions of the parameter of the model (amplitude, mean and relaxation time) from a given experimental time-series (PRE, 2019,110, 062142). We are currently working on applying these techniques to brain functional magnetic resonance measurements to discovery and characterize functional brain circuits.
Education
- Postdoc – National Institutes of Health, Bethesda MD, 1994-1998.
- Ph.D. - Biophysics, Technical University München, Germany, 1993.
- Diploma. - Physics, Technical University München, Germany, 1990
Academic Appointments
- 2008 - current: Associate Professor, Department of Biomedical Engineering, Stony Brook University, Stony Brook, New York
- 2004 - 2008: Assistant Professor, Department of Biomedical Engineering, Stony Brook University, Stony Brook, New York.
- 1998-2004: Assistant Professor, Department of Polymer Science and Engineering, UMass Amherst, Amherst, MA.
Honors
- 1994-1998: Fogarty Visiting Fellowship Award, National Institutes of Health.
- 1994-1996: Research Fellowship, Deutsche Forschungsgemeinschaft. Germany.
- 1995: Schlossmann Award, Max-Planck Society, Germany.
- 1999: GenCorp Signature Award
- 2000-2005: NSF CAREER Award (Condensed Matter Physics)
- 2003: Dillon Medal (American Physical Society)
- 2020: Weston Visiting Professorship at the Weizmann Institute in Rehovot, Israel
- 2020: Fellow of the National Academy of Inventors
Publications
Click here to search Helmut H. Strey's Google Scholar listing.
Patents
- H. Strey, R. Kimmerling, T. Bakowski, U.S. patents #9625454 and #15908453 “Rapid and continuous analyte processing in droplet microfluidic devices”
- L. Mujica-Parodi, H. Strey, D. DeDora, US Patent App. 15/739,259 “Dynamic phantom for functional magnetic imaging”
Courses Taught
- BME 440/441 – Senior Design in Biomedical Engineering (SBU)
- BME 502 – Advanced Numerical Methods in Biomedical Engineering (SBU)
- PBQ 558 - Quantitative Biology (cotaught with Laufer faculty) (SBU)
- BME 572 – Biomolecular Analysis (SBU)
- BME 573 – iPhone programming for Biomedical Applications (SBU)