J. Peter Gergen, PhD
Research in the Gergen laboratory uses the developmental genetic framework of the Drosophila embryo to investigate the regulation of gene expression, with a primary focus on the Runt transcription factor. Runt is the founding member of the Runx family of transcriptional regulators. Runx proteins are present in all animal species examined, but not in plant or microbial eukaryotic systems. Runt is best characterized for its role in the Drosophila embryo where it has vital roles in several pathways, including sex determination, segmentation, and neurogenesis . Mutations in all three human Runx genes are associated with genetic disease, and targeted mutagenesis experiments in the mouse indicate that these genes have vital roles in several pathways, including hematopoeisis, neurogenesis and osteogenesis A unifying aspect of Runx function in these many different pathways is a role in cell fate specification. An intriguing aspect of regulation by Runt and the vertebrate Runx proteins is that they function both as transcriptional activators and repressors, depending both on the specific target gene and the developmental context. Indeed, this context-dependence is central to the Runt-dependent regulation of the segment-polarity genes during segmentation.
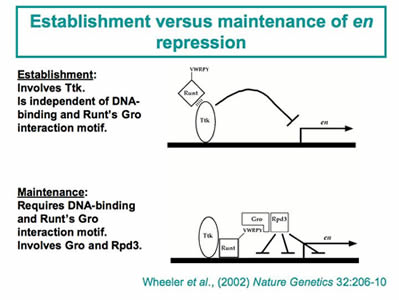
Our recent work has taken advantage of the differential sensitivity of different segment-polarity gene targets to quantitative manipulations in Runt activity. Runt is a potent repressor of the segment-polarity geneengrailed. We conducted a genetic screen to identify factors that contribute to this Runt-dependent repression. Further characterization of the factors identified in this screen revealed two distinct steps in en repression: 1) an establishment phase that involves a DNA-binding protein encoded by the tramtrack gene, and 2) a maintenance phase that involves the co-repressor Groucho and the Rpd3 histone de-acetylase. In vivo structure-function studies on Runt confirmed the functional distinction between establishment and maintenance and further revealed the surprising result that DNA-binding defective forms of Runt retain the ability to regulate a subset of gene targets. This genetic screen identified other factors that contribute to Runt’s potency, and ongoing work aims to further elucidate the roles of these factors in Runt-dependent regulation.
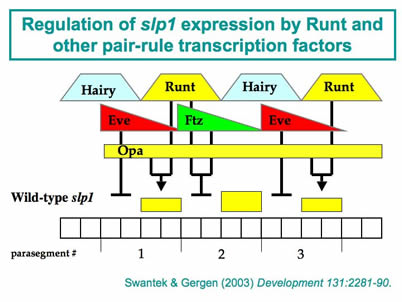
A second major project takes advantage of several attributes of the segment-polarity target gene sloppy-paired1 (slp1). One key advantage is the simple combinatorial rules that are responsible for modulating Runt’s activity as both an activator and a repressor of slp1expression. Indeed, using straightforward genetic manipulations it is possible to achieve uniform, physiologically relevant Runt-dependent activation or repression of slp1 in all somatic cells of a late blastoderm stage embryo. These embryos provide a platform for biochemical studies on the in vivoprotein-DNA interactions associated with slp1 repression. The results indicate that the initial establishment of repression does not involve changes in chromatin re-modeling or modification, nor the assembly of an initiation complex at the slp1 promoter, but is due instead to developmentally regulated transcriptional elongation. Ongoing studies are aimed at identifying the specific transcriptional step that is sensitive to regulation and characterizing the functional contributions of the factor(s) that are involved. This work is complemented by studies on the slp1 cis-regulatory region that investigate the quantitative contributions of different cis-regulatory DNA elements to developmentally regulated transcriptional elongation.
SUNY Distinguished Service Professor
Center for Developmental Genetics CMM/BLL 112
Stony Brook University
Stony Brook, NY 11794-5140
Office: 631-632-1190
Lab: 631-632-9031
Fax: 631-632-8575
john.peter.gergen@stonybrook.edu